Hanage, W. Microbiology: Microbiome science needs a healthy dose of scepticism.Nature 512, 247–248 (2014). https://doi.org/10.1038/512247a
Explorations of how the microscopic communities that inhabit the human body might contribute to health or disease have moved from obscure to ubiquitous. Over the past five years, studies have linked our microbial settlers to conditions as diverse as autism, cancer and diabetes.
This excitement has infected the public imagination. 'We Are Our Bacteria', proclaimed one headline in The New York Times. Some scientists have asserted that antibiotics are causing a great 'extinction' of the microbiome, with dire consequences for human health1. Companies offer personalized analysis of the microbial content of faecal samples, promising consumers enlightening information. Separate analyses from the same person can, however, vary considerably, even from the same stool sample. Faecal transplants have been proposed — some more sensible than others — for conditions ranging from diabetes to Alzheimer's disease. With how-to instructions proliferating online, desperate patients must be warned not to attempt these risky procedures on themselves.
Microbiomics risks being drowned in a tsunami of its own hype. Jonathan Eisen, a microbiologist and blogger at the University of California, Davis, bestows awards for “overselling the microbiome”; he finds no shortage of worthy candidates.
Previous 'omics' fields have faltered after murky work slowed progress2. Technological advances that allowed researchers to catalogue proteins, metabolites, genetic variants and gene activity led to a spate of associations between molecular states and health conditions. But painstaking further work dampened early excitement. Most initial connections were found to be spurious or, at best, more complicated than originally believed.
The history of science is replete with examples of exciting new fields that promised a gold rush of medicines and health insights but required scepticism and years of slogging to deliver even partially. As such, the criteria for robust microbiome science are instructive for all researchers. As excitement over the microbiome has filtered beyond academic circles, the potential mischief wrought by misunderstanding encompasses journalists, funding bodies and the public.
Crucial questions
Here are five questions that anyone conducting or evaluating this research should ask to keep from getting carried away by hype.
Can experiments detect differences that matter? Profiling a microbiome could produce a catalogue at the level of phyla, species or genes. Much work relies on analysis of 16S rRNA, an ancient gene that tolerates little variation and so is reliably found across the bacterial kingdom. But this allows only a coarse sorting. For example, microbiomes associated with obesity have been distinguished by different ratios of bacterial phyla, which encompass a staggering range of diversity. If this criterion were used to characterize animal communities, an aviary of 100 birds and 25 snails would be considered identical to an aquarium with 8 fish and 2 squid, because each has four times as many vertebrates as molluscs. Even within a single species, strains often differ greatly in the genes they contain.
Press officers must stop exaggerating results, and journalists must stop swallowing them whole.
Modern technology now allows for finer distinctions: we can study more genes in a sample, an ability that may enable us to decipher 'metabolic networks' revealing the biochemical reactions that a microbiome can perform. This kind of analysis could identify gene combinations, potentially from multiple species across a microbial community, that affect health for good or ill. However, pinning an outcome to any particular entity is likely to be hard unless the networks are already well characterized.
To take a simple example from a single bacterial species, we could show that vaccination eliminated 30% of known pneumococcal strains in a human population — but only because we knew in advance to focus on the genes targeted by the vaccine3. Our ability to identify functional differences in closely related genes is rarely sophisticated enough to pull out important genes or networks if we do not know what to look for in the first place. Moreover, genomes are littered with clues both true and false, such as 'hypothetical proteins' and genes that are understood poorly or not at all, but could make for important differences in what metabolic networks do.
We need to be able to identify functional differences in closely related genes from sequence alone. Until then, we must remember that apparent similarities might cloak important differences.
Does the study show causation or just correlation? A separate question is raised when distinct microbiomes can be identified and associated with diseases or other conditions. Then we are left with the chestnut of causes and correlates. Sometimes, a particular microbiome found in association with disease will be merely a bystander4.
A 2012 article comparing the gut microbiomes of old people living in care homes with those of old people living in the community found distinct microbiomes that correlated with multiple scores of frailty5. After accounting for some potentially confounding factors, the authors proposed a causal relationship: diet altered the microbiome, which in turn altered health. This explanation fits the data, but the reverse causality — the potential for poor health to alter the gut microbiome — was not explored. Frailer people probably have less active immune systems and differences in digestion (such as the time required for food to pass through the stomach and intestines) — factors that could change the microbiome. This work is not the only example of this sort of confusion.
What is the mechanism? All scientists are taught the catechism that correlation is not causation, but correlation almost always implies some sort of causal relationship. We just don't know what it is. We must determine it with careful experiments.
In the past three or four years, studies have advanced from characterizing a broad community of mainly unculturable microbes to identifying functional elements, individual taxa or particular properties. We can now design experiments to precisely define actions of components of the microbiome6, for example by reconstituting communities but leaving out specific taxa, or by precisely measuring the biochemical activity of an experimental microbiome in an 'organ on a chip'7. A return to a reductionist approach is essential if we are to pinpoint both whether the microbiome affects human health, and exactly how it does so.
How much do experiments reflect reality? Even if the microbiome can have an experimental effect, it may not be an important cause of the symptoms seen in ill people.
Much work has addressed the role that gut flora have in obesity, and several studies have found associations between the gut microbiome and weight gain8. To assess whether this association was cause or consequence, researchers collected gut-microbiome samples from human twins (one obese, one not) and introduced the microbiota to mice. Mice previously colonized with an 'obese' microbiome lost weight when supplied with a 'lean microbiome', but only if also fed a normal or low-fat diet. Diet alone had little effect9. Although this elegantly controlled experiment suggests great potential for the microbiome and related therapies to affect health, it also shows the microbiome's limits: the effect was dependent on other factors, in this case diet.
Microbiome studies often rely on germ-free mice. These animals allow researchers to readily introduce an experimental microbiota. But they do not represent the animals' natural state and are typically unhealthy owing to the lack of a microbiome. So results may not predict responses in animals with flourishing microbiomes. Mice and their microbiomes are also adapted to a rather different niche from humans, so results may not be generalizable.
Could anything else explain the results? There are good reasons to think that bacteria influence us in a host of ways. But there are many other — possibly more important — influences, such as diet in the earlier example. Whenever a study links a microbiome to a disease, wise critics should ask whether other contributors to disease are considered, compared and reported.
The hype surrounding microbiome research is dangerous, for individuals who might make ill-informed decisions, and for the scientific enterprise, which needs to develop better experimental methods to generate hypotheses and evaluate conclusions. Funding agencies must not let their priorities be distorted by the buzz around the field, but look dispassionately at the data. Press officers must stop exaggerating results, and journalists must stop swallowing them whole. In pre-scientific times when something happened that people did not understand, they blamed it on spirits. We must resist the urge to transform our microbial passengers into modern-day phantoms.
----------------------------------------------------------
Editorial
Hype or hope?
Nat Rev Microbiol17, 717 (2019). https://doi.org/10.1038/s41579-019-0283-5
Microbiome research has attracted considerable attention, partially because of the potential to manipulate the microbiome for human health. To fulfil this promise, tractable methods and cautious interpretation of results are needed.
The microbiota is booming — the number of studies mentioning ‘microbiome’ or ‘microbiota’ in their title or abstract grew from 11 in 1980 to over 13,000 in 2018. Almost any human disease you can think of has proposed links with the microbiome: inflammatory bowel disease, cancer, diabetes, obesity, atherosclerosis, fatty liver disease, malnutrition, autism, Alzheimer disease, depression, autoimmunity, asthma and on goes the list. Do you want to run faster, sleep better, live longer, or have more friends? There is bound to be a claim for a ‘microbiota fix’. An array of microbiome-based therapeutics are on the market or in development, ranging from prebioctics, probiotics, postbiotics, poop pills and biologics that target the gut microbiota to probiotic skin care and vaginal microbiota transplants. Furthermore, various companies offer testing of your (or your dog’s) gut microbiome and then make personalized diet and lifestyle recommendations and some even sell supplements. And for the so inclined there are now even smart toilets that send faecal bacterial counts directly to your smartphone. As a microbiologist, one cannot help but think of some of these developments as the Wild West of microbiome science.
However, it is clear that we live in close association with our microbiota and their functions are manifold. Furthermore, many human diseases are complex and for some, current treatments or preventative measures are insufficient and targeting the microbiota could be a potential way to deal with some of these challenges. The prime example is Clostridioides difficile infection, which can be efficiently treated with faecal microbiota transplantation. Owing to technological and analytical advances, our knowledge of microbiota diversity and functions and their roles in disease is rapidly increasing, and efforts are under way to go beyond disease associations and to translate this knowledge into therapeutics and applications. In large part, successful translation relies on tractable methods and systems that enable systematic and controlled testing of hypotheses and interventions. To highlight the latest developments in microbiome tractability and translation, we have put together a special Focus issue on these topics.
successful translation relies on tractable methods and systems that enable systematic and controlled testing
Models are essential tools to understand disease mechanisms and the specific molecular links that determine host–microbiota associations and interactions. In recent years, microbiome research has increasingly expanded to the use of simple animal models such as flies, zebrafish and worms. Angela Douglas argues that these simple models are cost-effective and time-efficient, and are particularly useful for large screens or multivariate experimental designs.
Once such a screen has identified a trait or relationship to target, microbiome engineering can be used to develop interventions. Chris Lawson, Trina McMahon and colleagues detail the design-build-test-learn cycle to efficiently engineer microbiomes for various applications. This approach has been successfully used in other fields such as metabolic engineering and provides a tractable path for microbiome translation.
One potential application is manipulating the human gut microbiota to treat or prevent diseases such as diabetes mellitus or cardiovascular disease. There is a clear link between nutrition, the gut microbiome and health. However, large individual differences make it difficult to predict the direction and strength of responses and thus to therapeutically manipulate this link. Eran Elinav and colleagues discuss the considerable variation in how individuals and their microbiome react to diet and how personalisation could optimize any therapeutic or preventative interventions that target this axis.
The human gut is a complex environment and Jeroen Raes and colleagues argue that a systems ecology perspective is needed to understand the gut microbiota. They provide an outline of how culturing and in vitro interaction experiments, combined with computer modelling, can help us to develop a functional and mechanistic understanding of the gut microbiota, which is the basis for successful translational projects.
In summary, advances in modelling, be it animal, in vitro or in vivo models, and ever increasing datasets raise great hope for microbiota translation and applications. In this context, a little bit of hype might not only have negative consequences. After all, the strong interest in microbiome science has led to funding initiatives and industry engagement (as reflected by predicted growth rates of the microbiome market of over 20% per year). It is time to further strengthen the scientific basis from which microbiome translation can grow.
----------------------------------------------------------
The human microbiome:
opportunity or hype?
Pedro M. Valencia, Magali Richard, Jesse Brock and Elsy Boglioli
Nat Rev Drug Discov16, 823–824 (2017). https://doi.org/10.1038/nrd.2017.154
Advances in the understanding of the gut microbiome - the complex ecosystem of microorganisms and their genes in the human digestive tract - have revealed its staggering scale and diversity. This has led to the recasting of the gut microbiome as a ‘new organ’ vital to human physiology that varies in composition between individuals, over an individual’s lifespan, and in diseases such as gastrointestinal disorders, infections, cancer, diabetes and obesity.
So, does the microbiome field provide viable opportunities for biopharma companies to develop innovative therapies, or has it been overhyped? Here, we argue that the opportunity is real, but that multiple questions remain related to the science, clinical development, regulation and business models for microbiome-based therapies.
Therapeutic approaches and investment
Microbiome-based therapeutic approaches and companies are spreading rapidly. From 2005 to 2015, an estimated US$1.6 billion was invested in microbiome-related companies. Human health care and nutrition represent the main fields of interest for microbiome investors, accounting for >75% of the 40 companies and ~85% of the investments. Other areas include agriculture and animal health. Compared with other ‘hot’ fields such as immunotherapy, the level of investment
is small, but the ramp-up speed is similar. Therapeutic approaches being explored include: whole microbiome transplants; bugs as drugs and probiotics; prebiotics and contrabiotics; host–microbiome interaction pathways and next-generation antibiotics.
Whole microbiome transplant (5% of total investment) is an intervention that introduces microbiota from a healthy donor into a patient. This approach has been successfully used to treat Clostridium difficile infections. Despite an impressive 90% cure rate, faecal microbiota transplantation (FMT) faces key challenges, including establishing donor stool banks and ensuring sample quality, procedure optimization, and transplant durability. A few companies, including Rebiotix and MaaT Pharma, are developing the next generation of FMT therapies.
‘Bugs as drugs’ and probiotics (43% of total investment) act on the microbiome by introducing known beneficial microbes into a patient or promoting their growth. Companies such as Seres Therapeutics and Vedanta Biosciences are focusing on these approaches. Seres is the most advanced so far, and despite a phase II trial failure last year in patients with C. difficile infection, it is preparing another phase II trial with an improved design. A novel approach, now being tested by Synlogic, is to engineer microbes that produce beneficial compounds after introduction into the gut. Evelo Biosciences is leading the field in oncology based on the correlations between the microbiome and the immune system in cancer patients.
Prebiotics and contrabiotics (33% of total investment) are compounds that favour or block particular microbiota populations and thus modify the microbiome composition and structure. Companies including Ritter Pharmaceuticals and Synthetic Biologics are actively exploring prebiotic or contrabiotic compounds for gastrointestinal indications.
Host–microbiome interaction pathways (16% of total investment) are being explored based on the analysis of metabolites produced by the microbiome. The goal is to restore activity of the microbiome by introducing compounds, typically produced through gut fermentation, that act on known pathways - known as ‘microbiome mimics’. Companies, including Enterome and Second Genome, are focusing on a functional, mechanism-of- action angle by directly targeting the pathways.
Next-generation antibiotics (7% of total investment) target drug-resistant microbial strains and provide alternatives that avoid major side effects on the microbiome. In addition to developing small molecules, several companies, including Spero Therapeutics and Epibiome, are focusing on novel technologies such as bacteriophages and targeted peptides. Others, such as Eligo Biosciences, are combining microbiome science and gene editing tools to develop genetically engineered bacteriophages.
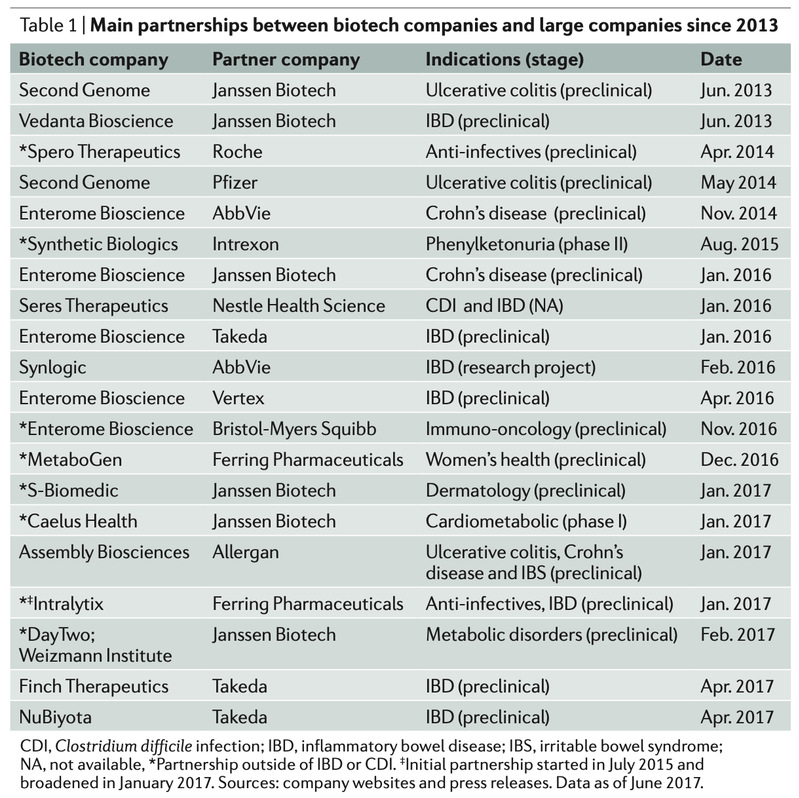
Despite the high level of activity by early entrants, significant opportunity remains across therapeutic areas and approaches. Large companies have been open to making initial investments in the field in the form of: product-related deals with biotech companies (TABLE 1); investing directly in biotech companies (such as Nestlé Health Science investing in Enterome and Seres Therapeutics); and investing in venture capital funds with a microbiome focus (such as Nestlé Health Science partnering with Flagship Pioneering, as well as Danone and Novartis in Seventure Partners’ microbiome-dedicated fund).
Questions for the path to market
There are several key development and commercialization questions about the manipulation of the microbiome as a new therapeutic modality. First, related to clinical development and regulation, how would clinical trial design need to evolve in terms of size, end points, comparators and outcomes? And would regulatory requirements need to adapt to the nuances of products acting on the microbiome? Natural changes to donor bacterial populations can affect the composition of bacterial isolations from day to day. To account for this, trials may require alternative comparators, other end points and additional outcomes.
Second, related to manufacturing, would new standards be required to ensure Good Manufacturing Practice product quality and safety (including reproducibility and scalability) of the samples? Bacteria are living organisms and are subject to changes in function and relative abundance between isolations and under different storage and culture conditions.
Third, related to market access and commercialization, will future microbiome-centred interventions primarily be preventive, curative or act as a supplement to other therapies? Will they be provided as a prescription drug or as part of a nutritional supplement? How will the delivery method affect the pricing and marketing strategies? While probiotics are already on
the market, treatments that radically alter the composition of the gut microbiome in an effort to reverse a disease state have yet to pass clinical trials. Developmental and regulatory complexity would influence how these products are priced and marketed.
Last, related to intellectual property (IP), are isolated bacterial strains patentable? Similar to the human genome, there is probably limited opportunity to patent products directly derived from nature, including bacteria and metabolites produced by bacteria. However, the unique combinations, genetic alterations, formulation methods and tools present an opportunity for companies to build IP.
Considerations for investment
Unlike other recent revolutions in biotechnology, the microbiome is not a new technology, but a new set of pathways associated with a ‘new organ’. Any therapeutic modality can be considered to target these pathways—small molecules, antibodies,proteins, gene editing, RNA technologies or even cells. So, the range of possible products is large, spanning many therapeutic areas.
The excitement around the microbiome is reminiscent of the progression of gene therapy. Compared with gene therapies, which must be precisely engineered at the molecular level, and in many cases delivered with precision to specific subcellular compartments, microbiome-based therapies may be easier to design. Microbial cells can be more easily reprogrammed, and at the organ level, the ecosystem displays a high degree of plasticity. However, there are other challenges.First, for many diseases, the causality of microbiome changes to the underlying disease mechanism has not been clearly elucidated (in contrast to gene therapies). Second, for therapies that depend on healthy donors, variation between donors may affect the overall efficacy. Finally, the high degree of plasticity may lead to unsustainable results.
With the accelerating pace in discovery and investment in the field, we expect that many of the unknowns will no longer be an issue in the coming years, and the first wave of therapies could be approved for gastrointestinal diseases and cancer, followed by a second wave in metabolic diseases (such as obesity and diabetes), and eventually for neurological diseases with the highest unmet needs. As in other fields that have profoundly changed human health, the journey is not likely to be a sprint, but rather a marathon.
