Recommended reading for bioinformatics V1
Keywords: bioinformatics, cardiometabolic diseases, microbiology, metagenomics, metabolomics, transcriptomics, omics integration, statistics, programming, NGS, bacterial evolution, machine learning
-----------------------------------------------------------------------------------------------------------
EMBL-EBI: online learning courses: https://www.ebi.ac.uk/training/
Recommended publications: see Labwiki Journal club list
Recommended book for bioinformatics and NGS:
Understanding Bioinformatics by Marketa Zvelebil;
Genomes 4 by TA Brown; Lewin's GENES XII;
Recommended protocols for common bioinformatical softwares or pipelines:
Nature Protocols
Pay attention to the following: Clustal W/X, Bowtie2, MUMmer, Hmmer, GeneMark, BEDTools, MOCAT2, QIIME, Mothur, FastQC, MEDUSA, OrthoMCL, MEGA, BLAST, BLAT, BWA, MAFFT, muscle, pfam_scan, star, HTSeq, bamtools, fastANI, GenomeTools, etc.
Recommended programing skills:
R
Python/Perl
Recommended books for microbiology:
Brock Biology of Microorganisms
Recommended books for biochemistry and Cell biology:
Lehninger Principles of Biochemistry;
Molecular Biology of the Cell
Recommended books for R:
Bioinformatics and Computational Biology Solutions Using R and Bioconductor;
Bioconductor case studies-Florian Hahne;
R Programming for Bioinformatics-Robert Gentleman;
R in a Nutshell-Joseph Adler
Recommended books for Statistics:
The Elements of Statistical Learning
Basics in maths:
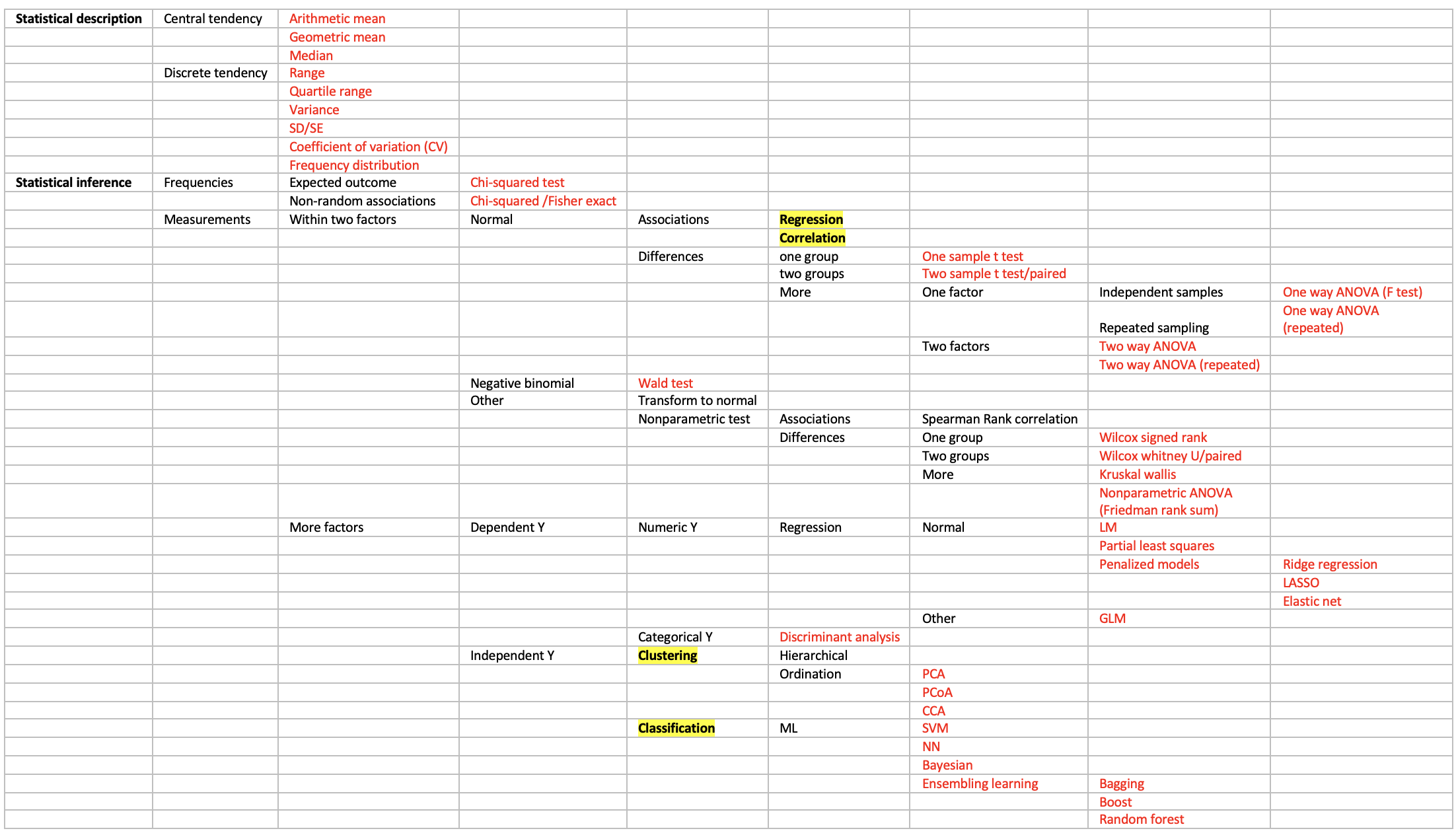
For reproducible research tools:
1) Conda for each project with all necessary softwares/Mamba (a better and faster version of conda written in C++);
2) snakemake;
3) Git/RMarkDown/Jupyter;
4) Docker
Refs for metagenomics analysis:
1) The CANOPY ref free method: Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 2014
2) The ref-based method: Metagenomic data utilization and analysis (MEDUSA) and construction of a global gut microbial gene catalogue. PLoS Comput Biol 2014 Vol. 10 Issue 7 Pages e1003706
3) MOCAT2: MOCAT2: a metagenomic assembly, annotation and profiling framework. Bioinformatics 2016
4) GEM for gut microbiome: High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol 2010 Vol. 28 Issue 9 Pages 977-82; An extended reconstruction of human gut microbiota metabolism of dietary compounds. Nat Commun 2021 Vol. 12 Issue 1 Pages 4728
5) PTRC: Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 2015 Vol. 349 Issue 6252 Pages 1101-6
Omics integration methods:
1) iCluster series: Pattern discovery and cancer gene identification in integrated cancer genomic data. Proc Natl Acad Sci U S A 2013 Vol. 110 Issue 11 Pages 4245-50
2) mixOmics: mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol 2017 Vol. 13 Issue 11 Pages e1005752
3) MOFA: Multi-Omics Factor Analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol 2018 Vol. 14 Issue 6 Pages e8124
4) PINS: A novel approach for data integration and disease subtyping. Genome Res 2017 Vol. 27 Issue 12 Pages 2025-2039
